Marine Microbiology and Generation of Natural Product Fraction Library
Over the past 30 years, the marine environment has emerged as a frontier in natural products discovery, with a focus on sponges, tunicates, and algae. More recently, the vast microbial diversity of the marine environment has been demonstrated as a promising resource of biological and chemical diversity. With this as a starting point, we have collected marine sediments from locations around the world, with an emphasis on samples from coastal Texas and South Carolina, as well as the Bahamas, Tonga, and Ecuador.
Using a combination of existing methods and new strategies, we have built a continually expanding collection of unique marine bacteria for our discovery efforts. We have had particular success using biochemical and environmental factors to enrich for novel actinomycete and a-proteobacteria diversity. Our approach aims to take advantage of the importance of bacterial small-molecule communication, such as quorum sensing, as a way to stimulate growth of specific types of bacteria.
Probing the Mechanism of Action of Natural Products
Historically, the most successful strategy for the discovery of novel cancer therapeutics has been the use of phenotypic screens in cell- or organism-based assays. A major advantage of phenotypic screens is that they can target any enzyme, receptor or other macromolecule in its biological context, without a priori knowledge of a target, thus allowing discovery of compounds that act on catalytic domains of enzymes, but also those that act through allosteric mechanisms or interrupt protein-protein interactions. Although the rate of discovery of interesting new small molecules is high using phenotypic approaches, the difficulty of target identification and verification is a bottleneck that is difficult to overcome.
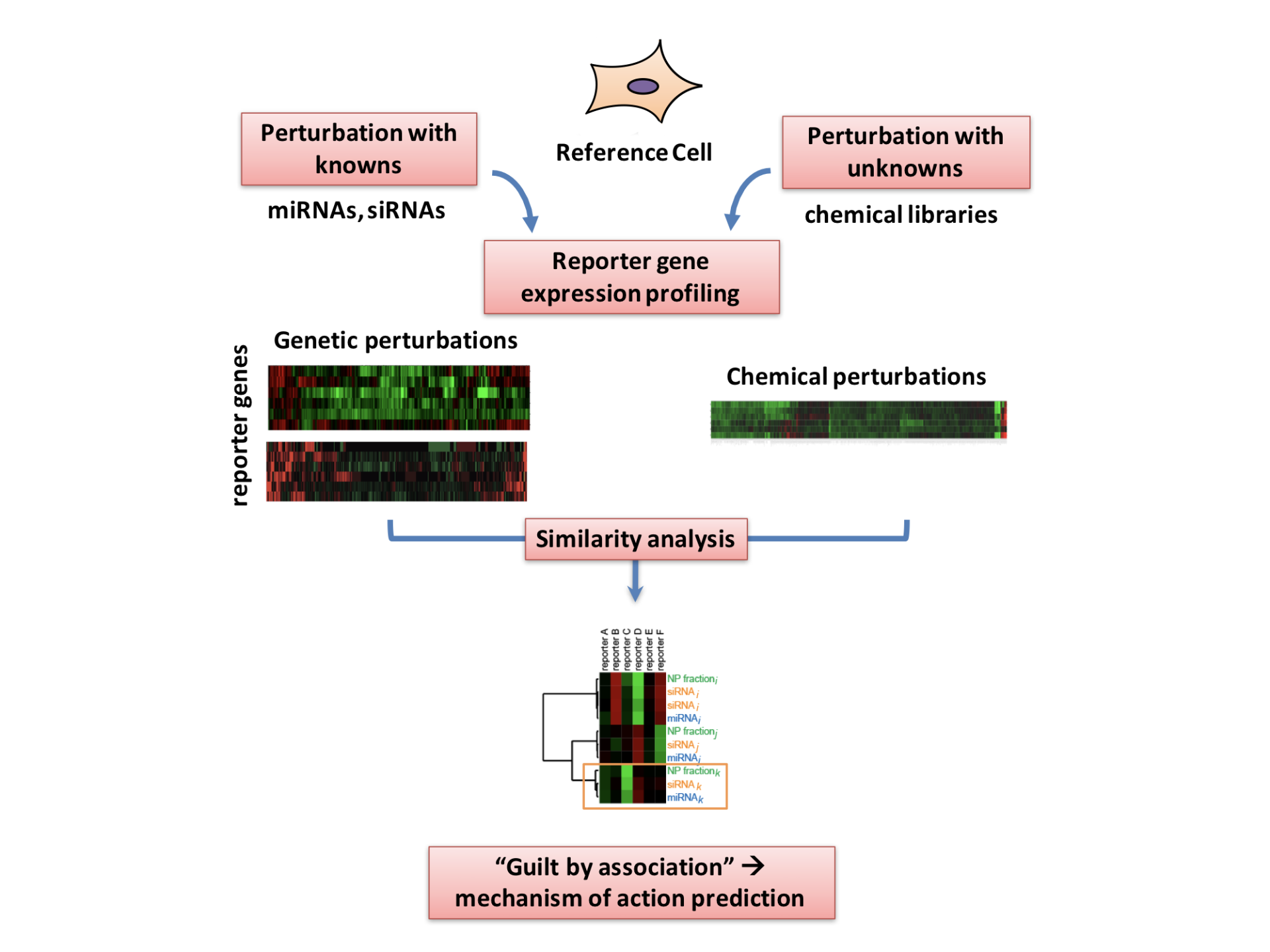
Working with the lab of Michael White, Ph.D., we have developed a method for the broad-scale classification of the biological activity of natural products in human cells, by employing an information-rich, high throughput, endogenous reporter gene expression platform that allows quantitative discrimination of concordant cellular responses to genetic and chemical perturbations. In a proof-of-concept, gene expression-driven functional signatures were employed as cross-platform phenotypic discriminators to link concordant cellular responses to 1124 genetic (siRNA, miRNA) and 1186 natural product perturbations, providing Functional Signature of Ontology (FUSION) maps, allowing us to predict the mechanism of action of natural products.
Integration of Natural Products and High-Content Phenotypic Screening
The intricate chemical structures of natural products have the advantage of optimization over evolutionary time scales for interaction with biological systems at sub-µM potency. A proven drug-screening paradigm for natural product collections has been the use of cell-based phenotypic screens, such as cytotoxicity, for selection of molecules with reasonable physiochemical properties. The development of high-content phenotypic screens, such as image-based and reporter-based, has opened up tremendous possibilities for the study of natural products. We have integrated our natural products fraction library, which consists of ~6,500 natural product fractions, with a number of high-content phenotypic screens taking place at the UCSC and UTSW high-throughput screening facilities.
Non-Small Cell Lung Cancer
One area of primary interest is identifying molecules with selective activity against non-small cell lung cancer (NSCLC), the most prevalent form of lung cancer. Working with scientists at UTSW Medical Center (John Minna, M.D. Professor in the Hamon Center for Therapeutic Oncology, Michael A. White, Ph.D., Professor of Cell Biology, and Michael Roth, Ph.D., Professor of Biochemistry), we are screening our natural products fraction library against a panel of 30 NSCLC cell lines to identify selective toxins. This project is part of the Cancer Target Discovery and Development (CTD2) network.
Development of New Methods for Structure Determination and Biocatalysis
New NMR Methods – MDEC
One of the remaining limitations of small molecule NMR is the ability to obtain relative configuration of small molecules. Recently, J-based configuration analysis has been applied to determine the relative stereochemistry of complex acyclic and macrocylic small molecules and natural products. The utility of J-based methods relies on the ability to measure discrete coupling constants between protons (JHH), which can be difficult in molecules with complex multiplets and significant signal overlap. We have developed a method that can simultaneously decouple multiple signals to greatly simplify signal overlap. This pulse can be integrated with other 1D experiments, such as 1D-TOCSY, to isolate individual signals in complex spectra.
Development of Novel Methods for Biotransformation
Biocatalysis has become an important component in the toolbox of the pharmaceutical and fine-chemical industries, with bioenzymes being used as reagents in multi-step synthetic processes. The biggest role for biocatalysis in the pharmaceutical sector lies on the chemo-, regio-, and stereoselectivity properties that biological systems can achieve.
As the pharmaceutical industry continues to expand its biocatalysis capabilities, there is still a limitation in the strategies to identify new reactions and enzymes. Discovery methods that can tap into the full potential of enzyme-catalyzed reactions, such as carbon bond-forming reactions, will expand the toolbox for this growing field. We have developed a biotransformation discovery platform that takes advantage of our marine-derived bacteria and the use of labeled substrates. The strategy that we developed utilizes the incorporation of a 13C label on the substrate, and allows 13C NMR spectroscopy to be used as a primary screening tool.